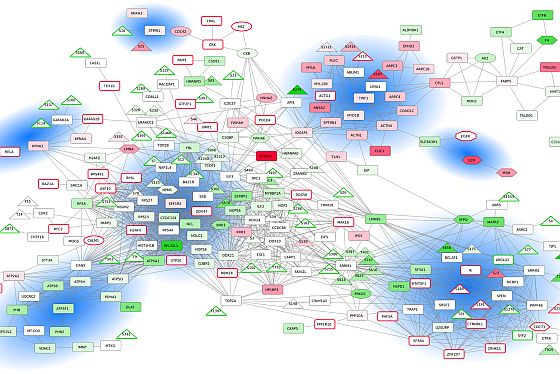
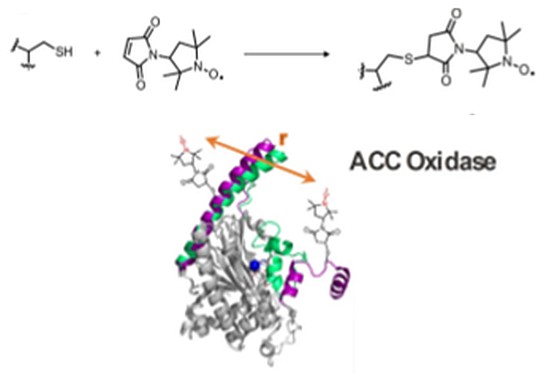
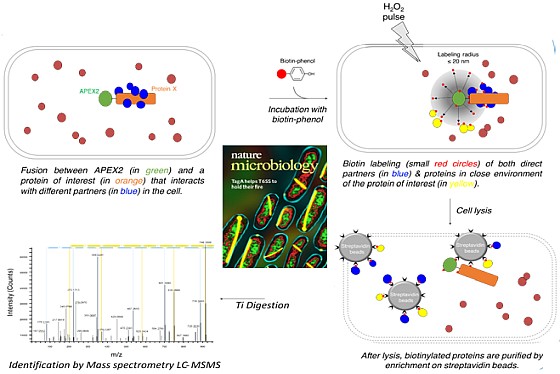
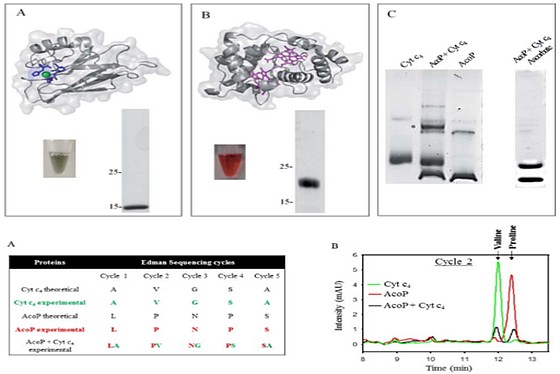
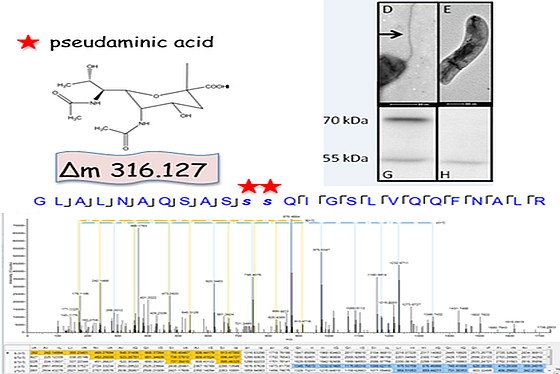
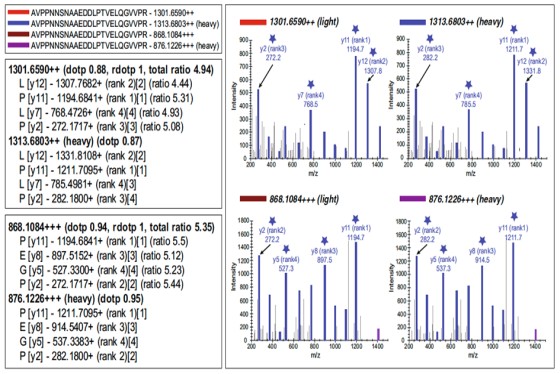
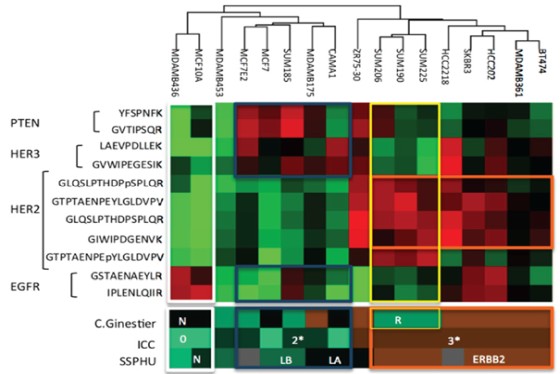
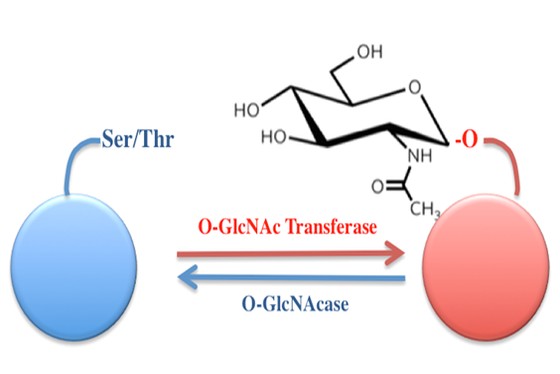
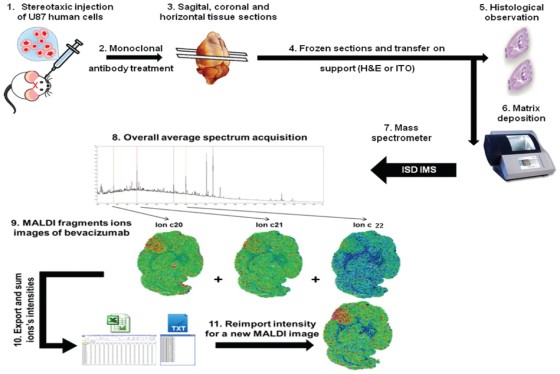
A regional multi-site platform
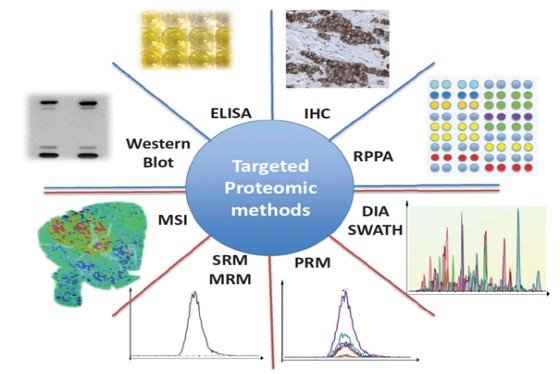
Marseille Protéomique (MaP) is a three-site proteomics facility, which besides mass spectrometry offers a wide panel of complementary techniques for protein identification and characterization, biomolecule quantification, biomarker discovery, binding studies imaging, structural proteomics and bioinformatics services. MaP is the main proteomics facility in the Région Sud Provence-Alpes-Côte d'Azur (PACA) region, which offers collaborative and fee-for-service opportunities for basic and clinical research but also for industry. MaP covers a large experimental field from oncology, immunology, and microbiology specific of each MaP component. PACA is in the third position in France for life science research and this dynamic scientific and medical environment brings to MaP numerous opportunities of collaborations at the regional level.
MaP was founded in 2007, accredited in 2008 by the Infrastructures Nationales en Biologie, Santé et Agronomie IBiSA and accredited in 2013 by Aix-Marseille Université. It is composed of three platforms located on three different sites close to the major research centres of the CNRS, INSERM and Aix-Marseille University, as well as the Institut Paoli Calmettes (Centre de lutte contre le cancer (CLCC)). Since 2024, MaP has been certified to ISO9001 (2015 version) and NFX50-900 (2016 version) standards by the certification agency LRQA.